Archive for the ‘Featured’ Category
Interactive Diagrams Using Inline JS+SVG+XHTML
>>> Attached: ( inline_svg_demo.xhtml — demo using the below technique for a silly drawing ) <<<
Compatibility: The following method doesn’t work for Internet Explorer 8, so if interoperability is really important to you, skip this series! Don’t worry though — Firefox 4.0, Internet Explorer 9 and WebKit (Safari / Chrome) will all natively support HTML5. HTML5 includes inline SVG for HTML so this ad hoc version can be retired when these three products take hold. The below method is compatible with Firefox 3.x, Opera 9.x and the current version of WebKit (Safari 5.x, Chrome 7.x). For display reasons, I’ll show off bitmap files inline while displaying inline SVG in linked pages.
Part One of Two
Part one is for everyone! I describe how to display an SVG inline on a webpage (XHTML) along with some JavaScript driven behaviour (clicking changes a displayed message and CSS defined colours).
Part two is for the bioinformaticians and phylogenists that survive and stick around — I utilize a parsed Newick format traversal to build up the drawing of a tree recursively — then I finish off by reintroducing JavaScript driven interactivity — tree nodes are highlighted via CSS when clicked, and information is displayed for the selected node (perhaps a multiple sequence alignment, a global alignment score, branch lengths — etc.).
( See part two here: Simple Interactive Phylogenetic Tree Sketches JS+SVG+XHTML )
Wait a sec — what do you mean by inline SVG?
Scalable Vector Graphics (SVG) is an XML vector image format. All browsers allow you to specify SVG files to be included into a webpage as an image. What we’ll be discussing here is how to include an SVG image as an XML element directly in a XHTML file — that is, the single webpage file will contain all of the markup for itself and its included SVG images without linking to any external files.
I won’t go into detail about how to make SVG images. You may create them with software like InkScape or by manual manipulation of the raw XML using a text or code editor (which I’ve done in this post). If you do end up using drawing software, simply copy out all of the XML elements with a text editor afterward and inject them into the XHTML page you create.
XHTML File Nuances
Include the following as a attribute of your html tag — this ensures that the browser reading your page knows which document type definition (DTD) to use.
<html xmlns="http://www.w3.org/1999/xhtml"> ... </html>
You must include your SVG with the following tags.
<svg xmlns="http://www.w3.org/2000/svg" width="300" height="300"
xmlns:xlink="http://www.w3.org/1999/xlink">
...
</svg>
In the above, we parameterize the “svg” tag with the correct xml namespace (xmlns) as an attribute. We specify the width and height of the image we want to inline and also specify that we want to be able to use hyperlinks within the SVG image with “xlink”. Notice that both namespace attributes are meant to enable a client browser to correctly render your page.
A Silly Drawing and Target Behaviour
In this post, we’ll be manipulating a picture of a silly face I’ve named Smile Again — if you can render the attached file, the face looks like the one to the right.
The eventual target behaviour we want is as follows. Clicking on different parts of Smile Again’s face causes (1) the displayed message to change and (2) the clicked part to change colours. Notice that I’ll be using JavaScript (JS) to perform both of these functions. We’ll get into the specifics soon — if you’re feeling up to it, you can always go to the attached demo xhtml file now and look at the source to reverse engineer the thing.
We can create this face manually with the following inline SVG code — we’ll start with pure SVG and worry about the JavaScript behaviour afterward.
<svg xmlns="http://www.w3.org/2000/svg" width="300" height="300" xmlns:xlink="http://www.w3.org/1999/xlink"> <circle cx="100" cy="100" r="40" style="fill:white;stroke:black;stroke-width:2" /> <circle cx="100" cy="100" r="20" style="fill:black;stroke:black;stroke-width:2" /> <circle cx="200" cy="100" r="60" style="fill:white;stroke:black;stroke-width:2" /> <circle cx="200" cy="100" r="10" style="fill:black;stroke:black;stroke-width:2" /> <path d="M60 200 C80 300 220 300 240 200 L60 200" style="fill:white;stroke:black;stroke-width:2" /> <path d="M100 235 L200 235" style="fill:white;stroke:black;stroke-width:2" /> <path d="M130 224 L130 246" style="fill:white;stroke:black;stroke-width:2" /> <path d="M150 220 L150 250" style="fill:white;stroke:black;stroke-width:2" /> <path d="M170 224 L170 246" style="fill:white;stroke:black;stroke-width:2" /> </svg>
Each of the XML elements above correspond to a different piece of the SVG image. The first four circles draw the eyes, next the complicated path draws the mouth and the four simpler paths draw the stitches of the teeth.
Changing the Displayed Message with JavaScript
Now that we’ve drawn Smile Again, we want to have it react to mouse clicks. I want to have both the clicked features highlight as well as the displayed message to change. This would be an important feature of actual practical diagrams. To make this task a bit easier to explain, I will break down the behaviour into smaller components and functions and build them back up as I go along. This will also make it a lot easier to read.
Let us focus on just the two circles that makes up Smile Again’s right eye (on the left side of the diagram).
We want our clicks to call a function — in order for that to happen, we need to use anchor tags (hyperlinks) within the SVG. This is made possible with our careful xmlns (namespace) attributes we declared earlier!
<svg xmlns="http://www.w3.org/2000/svg" width="300" height="300"
xmlns:xlink="http://www.w3.org/1999/xlink">
<a xlink:href="#" onclick="javascript:change_thoughts('Oww! My right cornea!')">
<circle cx="100" cy="100" r="40"
style="fill:white;stroke:black;stroke-width:2" />
</a>
<a xlink:href="#" onclick="javascript:change_thoughts('Ouch! My right iris!')">
<circle cx="100" cy="100" r="20"
style="fill:black;stroke:black;stroke-width:2" />
</a>
...
</svg>
Notice that we’ve now enclosed entire clickable elements in anchor tags and have used the “xlink:href” hyperlink attribute. In reality, we can have these links point to actual other webpages. Instead, these point to the same page and call the JavaScript function to change the text in the description called “change_thoughts”. In this case, the output is an expression of pain after being poked in the eye (“Oww! My right cornea!”). We can now define the “change_thoughts” function and also the place where we want our messages to appear.
<head>
...
function change_thoughts(say_this) {
document.getElementById("thoughts").innerHTML = "\"" + say_this + "\""
}
...
</head>
<body>
...
<h2 id = "thoughts" style="font-family:monospace;">
</h2>
...
</body>
The “change_thoughts” function can go in the head or somewhere early in the body of the XHTML. It doesn’t really matter as long as it occurs before the SVG. This function gets an element with the id “thoughts” and changes the HTML inside of it. In the above, I add a pair of double quotes to the string to show. The element with the id “thoughts” is a h2 element — this really could been any valid HTML element. You do however have to place the “thoughts” element somewhere after you declare the “change_thoughts” function.
Changing the Highlighting with JavaScript and CSS
The last thing we want to do is to change the highlighting by making our JavaScript function touch the style (CSS) of the inline SVG element that was clicked. We could use a separate CSS file, but in the spirit of making everything one inlined file, the CSS is specified with the “style” html attribute.
Again, we’ll be looking only at the two circles corresponding to Smile Again’s right eye (I’ve changed the way the lines are broken in the code listing below so that it will actually fit on this page).
<a xlink:href="#"
onclick=
"javascript:change_thoughts('Oww! My right cornea!','right_cornea', '#F00')">
<circle cx="100" cy="100" r="40"
style="fill:white;stroke:black;stroke-width:2"
id="right_cornea" />
</a>
<a xlink:href="#"
onclick=
"javascript:change_thoughts('Ouch! My right iris!', 'right_iris', '#00F')">
<circle cx="100" cy="100" r="20"
style="fill:black;stroke:black;stroke-width:2"
id="right_iris" />
</a>
Here are our two changes. First — each of the SVG elements now have a unique id (“right_cornea”, “right_iris”) so that we can use JS to manipulate each shape independently. Second — we’re now calling a new version of “change_thoughts” — this time, we’re giving the function the id of the shape to highlight and the colour (in hexadecimal) to highlight it with.
Finally, I can show you the new version of “change_thoughts” which will accept the new arguments and change the colours of the shapes.
function change_thoughts(say_this, highlight_this, colour) {
// Say this ...
document.getElementById("thoughts").innerHTML = "\"" + say_this + "\""
// Highlight this ...
var change = document.getElementById(highlight_this)
change.setAttribute("style",
"fill:" + colour + ";stroke:black;stroke-width:5")
}
In the above, we’ve added the code after “// Highlight this …”. We select the element “highlight_this” — remember, the id and colour are given by the function called with mouse clicks in the SVG. We then change the style by overwriting the inline CSS with the new specified fill colour (and a fat black stroke).
We now have a problem — each new mouseclick on a different shape will cause that new shape to highlight. The previous highlighted shape doesn’t automatically revert, and eventually all of the shapes are highlighted. What we need to do now is to improve the “change_thoughts” function one last time with some way to “undo” the previous change, so that new calls to this function will not only highlight the new shape, but will also remove the highlight from the previous shape.
Finishing with an Undo Object to remove previous changes
In the following version, we add an object to behave like a hash in the JavaScript. This hash will take html id attributes as keys, and the previous style as values. This behaves like an “undo” stack since we apply the styles to the elements with the corresponding id before we commit the changes to the new shape.
var _undo_render_hash = new Object
function change_thoughts(say_this, highlight_this, colour) {
// Say this ...
document.getElementById("thoughts").innerHTML = "\"" + say_this + "\""
// Undo previous highlight ...
for(var i in _undo_render_hash) {
document.getElementById(i).setAttribute("style", _undo_render_hash[i])
}
// Reset undo stack ...
_undo_render_hash = new Object
// Highlight this ...
var change = document.getElementById(highlight_this)
_undo_render_hash[highlight_this] = change.getAttribute("style")
change.setAttribute("style",
"fill:" + colour + ";stroke:black;stroke-width:5")
}
Walking through the code, we start with a blank object as the undo stack — this is OK. On the first call of this function, since the undo object is empty, simply nothing is done before the new shape is highlighted. After the first call, all of the changes that were committed in the last call of the function are undone as their original styles are reapplied. We just add the next object to change to the undo object before applying changes to it.
The undo object is treated like a hash here, and in fact, we iterate all of its elements even though we only expect to have captured one id in the last call of the function. This means we can use this undo technique for functions that will modify many shapes with many changes since each of those modifications is iteratively undone in the next call. We must of course ensure that we save the original style of each of these shapes.
Next
At some point, I’ll want to revisit this and combine some of the other JavaScript stuff I’ve shown in this blog before. Displaying a rendered clickable phylogenetic tree is a task I figured out since I needed to quickly visualize some data for my team in my thesis work. Since I’ve already covered parsing a newick tree in this blog, it makes sense to complete the discussion with how I ended up with my phylogeny visualizer.
The Null Coalescing Operator (C#, Ruby, JS, Python)
Null coalescence allows you to specify what a statement should evaluate to instead of evaluating to null. It is useful because it allows you to specify that an expression should be substituted with some semantic default instead of defaulting on some semantic null (such as null, None or nil). Here is the syntax and behaviour in four languages I use often — C#, Ruby, JavaScript and Python.
C#
Null coalescencing in C# is very straight forward since it will only ever accept first class objects of the same type (or null) as its operator’s arguments. This restriction is one that exists at compile time; it will refuse to compile if it is asked to compare primitives, or objects of differing types (unless they’re properly cast).
Syntax:
<expression> ?? <expression>
(The usual rules apply regarding nesting expressions, the use of semi-colons in complete statements etc..)
A few examples:
DummyNode a = null; DummyNode b = new DummyNode(); DummyNode c = new DummyNode(); return a ?? b; // returns b return b ?? a; // still returns b DummyNode z = a ?? b; // z gets b return a ?? new DummyNode(); // returns a new dummy node return null ?? a ?? null; // this code has no choice but to return null return a ?? b ?? c; // returns b -- the first item in the chain that wasn't null
No, you’d never really have a bunch of return statements in a row like that — they’re only there to demonstrate what you should expect.
Ruby, Python and Javascript
These languages are less straight forward (i.e. possess picky nuances) since they are happy to evaluate any objects of any class with their coalescing operators (including emulated primitives). These languages however disagree about what the notion of null should be when it comes to numbers, strings, booleans and empty collections; adding to the importance of testing your code!
Syntax for Ruby, Javascript:
<expression> || <expression>
Syntax for Ruby, Python:
<expression> or <expression>
(Ruby is operator greedy :P.)
The use of null coalescence in these languages are the same as they are in C# in that you may nest coalescing expressions as function arguments, use them in return statements, you may chain them together, put in an object constructor as a right-operand expression etc.; the difference is in what Ruby, Python or Javascript will coalesce given a left-expression operand. The below table summarizes what left-expression operand will cause the statement to coalesce into the right-expression operand (i.e. what the language considers to be ‘null’-ish in this use).
| Expression as a left-operand | Does this coalesce in Ruby? | Does this coalesce in Python? | Does this coalesce in JavaScript? |
| nil / None / null | Yes | Yes | Yes |
| [] | No | Yes | No |
| {} | No | Yes | n/a* |
| 0 | No | Yes | Yes |
| 0.0 | No | Yes | Yes |
| “” | No | Yes | Yes |
| ” | No | Yes | Yes |
| false / False / false |
Yes | Yes | Yes |
*Note that in JavaScript, you’d probably want to use an Object instance as an associative array (hash) so that the field names are the keys and the field values are the associated values — doing so means that you can never have a null associative array.
Contrast the above table to what C# will coalesce: strictly “null” objects only.
The null coalescing operator makes me happy. Hopefully it’ll make you happy too.
Parsing a Newick Tree with recursive descent
>>> Attached: ( parse_newick.js – implementation in JS | index.html – example use ) <<<
Update: Fixed parse_newick.js source code that caused a field to be mislabelled in index.html (2010 Dec. 19)
This method of parsing works for any language that allows recursive functions — basically any of the curly bracket languages (C#, C, Java) and the fast-development languages (Ruby, JavaScript, Python) can do this. This particular example will include JavaScript code which you can take and do whatever you want with — recursive descent parsing is a well-known and solved problem, so it’s no skin off my back. Before we begin, I should probably explain what a tree in Newick format is.
What exactly is the Newick format?
Remember back in second year CS when your data structures / abstract data types / algorithms instructor said you can represent any tree as a traversal? Well, if not — don’t fret, it’s not hard to get the hang of.
A traversal is a finite, deterministic representation of a tree which can be thought of as a road map of how to visit each node of the tree exactly once. The phylogeny / bioinformatics people will know each of these nodes as either taxa / sequences (the leaves) or profiles (the internal nodes — i.e. not the leaves). A tree in Newick format is exactly that — a traversal. More specifically, it is a depth-first post-order traversal. When traversing a tree, we have a few options about how we want to visit the nodes — depth-first post-order means we want to start at the left-most, deepest leaf, and recursively visit all of the nodes before we end at the root.
A tree expressed in Newick format describes a tree as an observer has seen it having visited each node in a depth-first post-order traversal.
A node in the Newick format can be…
- A leaf in the form: “<name>:<branch_length>”
- e.g. “lcl|86165:-0“
- e.g. “gi|15072471:0.00759886“
- e.g. “gi|221105210:0.00759886“
- Or a node in the form “(<node>,<node>):<branch_length>”
- This is where it gets tricky and recursive — the “node” item above can be another node, or a leaf — when a node is composed of at least another node, we call that a recursive case; when a node is composed of only leaves, we call that a base case. We make this clearer in the next section.
- Or a node in the form “(<node>,<node>);”
- The semi-colon at the end means that we have finished describing the tree. There is no branch length as the root of the tree is not connected to a parent.
Understanding recursion in a Newick Tree
Base case e.g.: The below node is composed of two leaves.
“(lcl|86165:-0,gi|87118305:-0):0.321158”
Recursive case e.g.: The below node is composed of a leaf on the left, and another node on the right; this right-node is composed of two leaves.
“(gi|71388322:0.0345038,(gi|221107809:0.00952396,gi|221101741:0.00952396):0.0249798):0.0111081“
If the names look a bit funny, that’s because these are sequences pulled out of a blastp search.
The Newick tree itself may have any number of nodes, the base case being a single node. The only restriction is that any node either is a leaf (has zero children) or must have two children (never a single child). These two children again are allowed to be either leaves or other nodes.
We would visualize the above Recursive case e.g. as follows.:
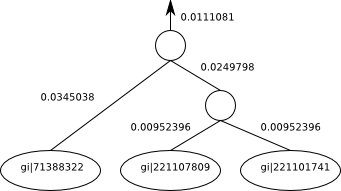
Where the numbers beside the edges are edge length and the names of the named taxa are indicated inside the circles (leaves). The unnamed internal nodes are the profiles that have been calculated with alignments. You’ll notice that there’s a branch length leading out of the root of this subtree into the rest of the tree (not shown). If this was a complete tree on its own, the root would have no branch length there and no arrow coming out of it.
Finally, Parsing!
Now that we have reviewed the depth-first post-order traversal, the Newick format and how to interpret it; we can move onto parsing these things. In parsing, we hope to create an in-memory structure that represents the tree. The parser I discuss assumes that the Newick format string already has had all newlines stripped, as it uses regular expressions. If your trees aren’t stored that way, just make sure that newlines are stripped before feeding it into my function. The in-memory object that’s created will have the following fields as a minimum.
- node_type $left – the left child of a node
- node_type $right – the right child of a node
- string_type $name – the name of a node (if it is a leaf)
- float_type $branch_length – the length of the branch coming out of the top of a given node
I also use the following two fields to keep track of a few house keeping items and to make life pleasant for future tasks with this in-memory structure.
- node_type $parent – the parent of a node
- integer_type $serial – the order in which this node occurs when parsed
Note that in future, a depth-first post-order traversal will yield the nodes in the exact order that they were read — so in reality “this.serial” isn’t really needed, but it’s nice to have for future functions you might write as a shorthand. Have you understood why these nodes would occur in the same order?
Each language has its own quirks. Python and JavaScript have the advantage that you can define a class and define its properties (fields) on the fly — meaning there’s less code to download and to write, which is particularly good for blogs 😛
Here’s pseudo-code for the recursive descent parsing function “tree_builder(<newick_string>)” — I don’t indicate where to strip away newlines because I assume they’re gone. I also don’t indicate where to strip away whitespace (I did previously, but that took up like half the pseudocode). So if there’s a place where you suspect whitespace can occur, strip it away and increment the cursor by the length of that whitespace.
Note: We are assuming that there are no newlines in the file — that is, the entire newick traversal is one string on a single long unbroken line.
// Note:
// Call tree_builder() with a "new" operator in JavaScript
// to allow it to use the 'this' keyword.
// In other languages,
// you may need to explicitly create a new object instead of using 'this'.
var count = 0; // can be either a global variable or a reference
// keeps track of how many nodes we've seen
var cursor = 0; // also either a global or a reference
// keeps track of where in the string we are
function tree_builder(newick_string) {
// Look for things relating to the left child of this node //
if $newick_string [ $cursor ] == "(" {
$cursor ++; // move cursor to position after "(".
if newick_string [ $cursor ] == "(" {
// Then the $left node is an internal node: requires recursion //
$this.left = new tree_builder(newick_string); // RECURSIVE CALL.
$this.left.parent = $this;
}
// try to find a valid $name next at the current cursor position //
if $name = $newick_string [ $cursor ] . regex match ("^[0-9A-Za-z_|]+") {
// Then the $left node is a leaf node: parse the leaf data //
$this.left = new Object;
$this.left.name = $name;
$this.left.serial = $count;
$count ++;
$this.left.parent = $this;
$cursor += length of $name;
// move cursor to position after matched name.
}
// notice: if no $name found, just skip to finding branch length.
}
if $newick_string [ $cursor ] == ":" {
// Expect left branch length after descending into the left child //
$cursor ++; // move cursor to position after the colon.
// look for the string representation of a floating point value "FPV"...
$branch_length_string =
$newick_string [ $cursor ] . regex match ("^[0-9.+eE-]+");
$this.$left.$score = $branch_length_string as a FPV;
$cursor += length of the string $branch_length_string;
// move cursor after FPV.
}
// Look for things related to the right node //
if $newick_string [ $cursor ] == "," {
$cursor ++; // move cursor after the comma
if newick_string [ $cursor ] == "(" {
// Then the $right node is an internal node: requires recursion //
$this.right = new tree_builder($newick_string); // RECURSIVE CALL.
$this.right.parent = $this;
}
// try to find a valid $name next at the current cursor position //
if $name = $newick_string [ $cursor ] . regex match ("^[0-9A-Za-z_|]+") {
// Then the $right node is a leaf node: parse the leaf data //
$this.right = new Object;
$this.right.name = $name;
$this.right.serial = $count;
$serial ++;
$this.right.parent = $this;
$cursor += $length of $name;
}
// again, accept if no $name found and move onto branch length.
}
// Now looking for the branch length ... //
if $newick_string at $cursor == ":" {
// Expect right branch length after descending into right child //
$cursor ++; // moving past the colon.
$branch_length_string =
$newick_string [ $cursor ] . regex match ("^[0-9.+eE-]+");
$this.right.score = $branch_length_string as a FPV;
$cursor += length of the string $branch_length_string;
}
if $newick_string at $cursor == ")" {
$cursor ++; // move past ")".
}
// Expect the branch length of $this node after analyzing both children //
if $newick_string at $cursor == ":" {
$cursor ++;
$branch_length_string =
$newick_string [ $cursor ] . regex match ("^[0-9.+eE-]+");
$this.score = $branch_length_string as a FPV;
$cursor += length of the string $branch_length_string;
}
if $newick_string at $cursor == ";" {
// This case only executes for the root node-- //
// --the very last node to finish parsing //
$cursor ++; // to account for the semicolon (optional, consistency)
$this.serial = $count; // no need to increment $count here
}
// Return to complete a recursive instance (internal) or base case (leaf) //
return this; // not needed if "new" operator was used all along
}
The implementation that’s included in the attached JavaScript does two things differently than the above. First, I don’t force the user to break encapsulation by declaring globals. Instead, I use an object called “ref” which has two fields, “ref.count” and “ref.cursor”. The name “ref” is chosen to remind us that its fields are passed by reference. This is possible because the object “ref” itself is passed by reference (I could have called it anything, but I chose a name that suited its purpose). Imagine trying to pass “count” and “cursor” in this recursive function without an enclosing object — what would happen instead is that the value wouldn’t be updated, instead the integer values would be copied and modified locally in the span of single function instances. The “ref” object is passed as an argument to the “_tree_builder(<ref>, <newick_string>)” function internally where it is instantiated if “null” is passed in the place of “ref”. The calling user doesn’t even need to know about it, as it’s all wrapped together in a pretty package and the user just calls “tree_builder(<newick_string>)” as before — without the need for globals. The enclosing function even does the work of instantiating the root node with the “new” keyword so that the end user doesn’t even need to (and shouldn’t!) say “new”.
Finally, there is a traversal function, and an expose function. The traversal function returns the node objects in an array following in the natural depth-first post-order described previously. The expose function is a debugging function which writes the output using “thoughts_write(<message>)” which wraps “document.write(<message>)”. You can change “thoughts_write(<message>)” to print output elsewhere using “document.getElementById(<some_identity>).innerHTML += (<message>)” where “some_identity” is the “id” of some element in your HTML. Without going into detail, the traversal function also uses an internal “ref” object which refers to the array as it is filled.
Some other things you can try…
The JavaScript recursive descent parser is actually a simplified version of something that I threw together earlier. The major difference is that I tag leaf nodes with fasta protein sequences by passing a reference to an associative array (or hash) as an argument to “tree_builder(<newick_string>, <fasta_hash>)”. Remember, in JavaScript, an associative array is an instance of an Object, not an instance of an Array. Using non-integer field names in an Array is actually not well defined but kind of sort of works semi-consistently between browsers (i.e. don’t try!), but that’s a story for another day. Whenever the name of a leaf is parsed, I would look it up in the hash and put it in a field either “this.left.fasta” or “this.right.fasta” thereby conferring to the leaf nodes of the in-memory structure the appropriate fasta protein sequence.
Lessons Learned
In putting this thing together, I managed to relearn JS, recursive descent parsing and making JS pass things by reference up and down a recursive stack of function calls. Andre Masella helpfully reminded me that I didn’t need to use any kind of BNF or LALR(1) parser — just an easy single-function parser would do. This is actually a port of a C# parser which does the same thing that I wrote in an earlier step for phylogenetic analysis — the difference being that C# offers a “ref” keyword that allows one to specify arguments in a function’s parameter list as emulated references (functionally equivalent to using C’s asterisk and ampersand convention though not operationally equivalent).
Happy parsing! The code presented should be easy enough to understand for you to port it into the language of your choice for whatever purposes you design.
An Old Physiology Project — Operation Spinny Chair :D
I discovered this ancient report in my repository about three months ago– I’ve finally decided to put it up because it made my day reading the abstract again. This is definitely one of my prouder albeit sillier projects from the days of my undergrad.
Independent Research Project Acute centripetal acceleration is correlated with increased heart rate and R-wave amplitude
Matthew Boyle, Bryan Chung, Eddie Ma
Abstract
In the present study, we set out to discover the correlation between the exposure of acute centripetal acceleration in human subjects and cardiovascular function across the following three dimensions: Heart rate, R-wave amplitude and QRS interval. This was accomplished by measuring the above properties via Lead II Bipolar ECG trace, after having spun the subject at 0.8 revolutions per second in an office chair for successively, 30, 60 and 90 seconds. It was determined that heart rate showed strong positive correlation (n = 3, average increase between trials of increasing duration, 3.2 beats per minute, s = 1.8). R-wave amplitude showed positive correlation in all subjects up until and including the 60 second trial. There was no systemic correlation between duration of spin and the length of the QRS interval in any of the subjects. The heart is therefore an important effector in response to centripetal acceleration in the human model.
Key Words: electrocardiogram, QRS interval, centripetal force, R-wave amplitude, spinning office chair.
[ Download this | PDF ].Thumbnails of pages 5, 7 and 11.



Squishy TIM Barrel Subunits
Again with the TIM Barrel pictures! Here’s some text about it from my notes…
1a5m (A Urease) is a really interesting protein– it consists of three subunits. Each subunit consists of three unique domains: a very squashed TIM Barrel, an alpha-alpha-alpha-beta-beta domain and a beta-beta-alpha-beta-beta domain. I’m not yet sure what to call little broken alpha helices that have less than two complete turns. The TIM Barrel (though exceedingly asymmetrical) will still be accounted for in the data to be analyzed. The TIM Barrel (566 amino acids) is the alpha subunit of each symmetrical subunit. The remaining two domains are the alpha and beta subunit though PDB is not clear which is which: they each weigh in at 100 and 101 amino acids. 1a5m is part of several solved urease structures in the PDB– the collection: {1A5K, 1A5L, 1A5M, 1A5N, 1A5O} are solved by Pearson et al. (1998).
References
Matthew A. Pearson, Ruth A. Schaller, Linda Overbye Michel, P. Andrew Karplus, and, Robert P. Hausinger (1998). Chemical Rescue of Klebsiella aerogenes Urease Variants Lacking the Carbamylated-Lysine Nickel Ligand. Biochemistry. 37(17):6214-20.
Squishy squishy shapes– the giant pink object in the next picture is actually three such TIM Barrels, each of which belongs to one of the three subunits.
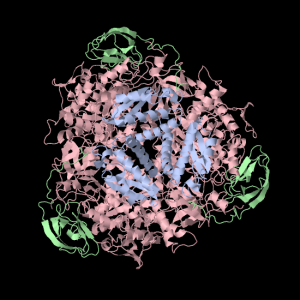
Each of the three subunits are shown separately below…
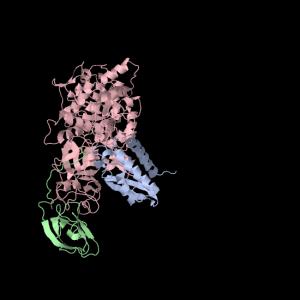 |
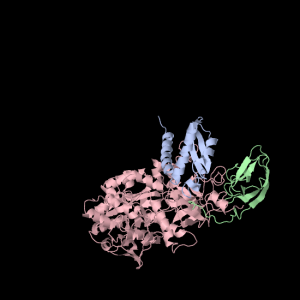 |
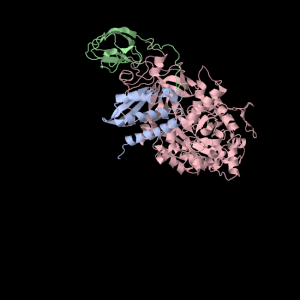 |
Aesthetically pleasing– these images were captured from the JMol output available from RCSB PDB.
New Diagram for MSc-X3 (math paper)
Brief: I’m particularly happy with this diagram… I had something along these lines in my head for a while, but I never could figure out how to draw it correctly. I never thought that simplifying it to three easy steps was the smarter thing to do.
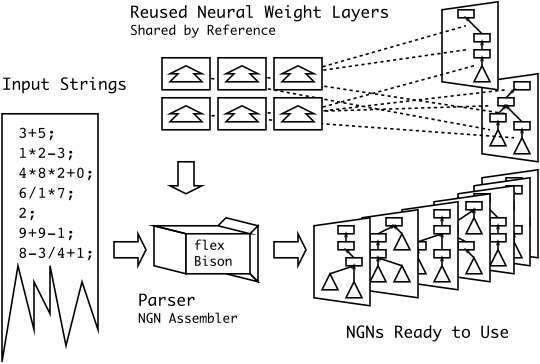
 Ed's Big Plans
Ed's Big Plans
