Archive for the ‘Stefan C. Kremer’ tag
New Diagram for MSc-X3 (math paper)
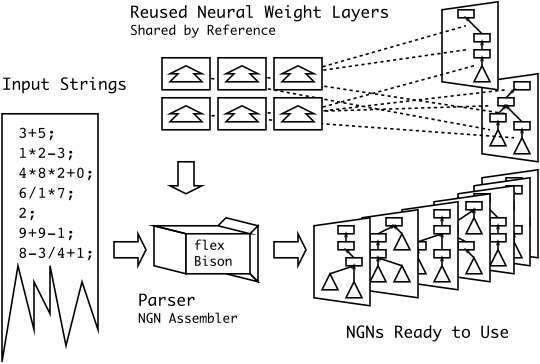
Brief: I’m particularly happy with this diagram… I had something along these lines in my head for a while, but I never could figure out how to draw it correctly. I never thought that simplifying it to three easy steps was the smarter thing to do.
 Ed's Big Plans
Ed's Big Plans