Archive for the ‘Academic Life’ Category
The Computational Bio & Chem Group (SOLVER Revisited)
A Revisitation
In the beginnings of the Winter semester, I had an idea to start up a undergraduate/graduate student group that would provide a scaffold for faster, computer assisted research.
The semester was simply too full for me to try and get it running at that time. I’m tempted to do some work on it later this semester, after I’ve gotten more of my thesis done.
The basic idea is as follows. Faculty and graduate students in the biology and chemistry departments often have the need to analyze data with some elegant computing. Whether this be as commonplace as hacking together an excel sheet to do work or learning some existing toolbox, or something slightly more in depth such as analysis in R, Python or PERL. Unfortunately, these research problems often fall by the wayside as the time commitment to learn a new software package or programming language is not trivial without a stronger computing background. Undergraduate students who are raised in the computer science environment, particularly with a bioinformatics interest have some knowledge of the research problem semantics as well as some knowledge of how to do the above analysis by using and coding software.
The “SOLVER” group would fill the gap by performing a matchmaking and coaching service. In my vision, SOLVER creates working teams of three to four– (1) one or two graduate students or faculty with a research problem, (2) one or two undergraduate students with some knowledge in bio / chem and some talent in computing, (3) an experienced coach that can recommend best practices so that the team has a good shot of solve the problem in a reasonable amount of time. In the end, the researcher gets help and a good chance at a working solution– they might even learn some programming; educational / professional / social connections are made; and the student gets an item to add to their resume. In reality, a particular research step would be executed as week-long blocks– whether this means one block or four blocks (one month) depends on the complexity of the task.
To do…
One stretch of work that I have to do is to determine the needs of the department. I wanted to do this in the middle of Winter semester but didn’t find it a priority. For this group to work, there must be research problems. Similarly, I need to determine the capabilities of potential undergraduate students. I’m learning quickly that the key here is to start small and think big (i.e. start with one group, then two, then learn about the administrative logistics, then deploy some progress tracking mechanism, then four groups and onward).
Retuning the image
The almost idealized description above suffers from a few logistical issues. First, I will now address the issue of publication credits. Because researchers must attribute tangible work (including written code and analyses), some graduate students may be hesitant to participate in the program. I don’t know if this will actually become a barrier however as a “tangible contributor” would never be the first nor the last name on a paper (this is true in all CS, Bio and Chem). Furthermore, a paper with more than one author demonstrates the ability to collaborate in a team, and fostering the experience of another student is part of our culture (e.g. co-op students etc.). That brings me to a second major issue. One of the frames that this group could find itself in is a means to circumvent or short-circuit the co-op student appointment process– a frame that I readily reject. In fact, I should hope that this group becomes a means to introduce new putative co-op students to their future advisers which may otherwise be overlooked for their differing background. Finally, there is the problem of attrition, whereby a group dwindles in size as members drop out. The only contract-based perks or penalties I can think of is really ties to the group itself (e.g. unsatisfactory work naturally results in a time out or withdrawal from the program). It is really the only tangible leverage we can offer at the outset– unsteady leverage at best (difficult to assume a reputation when none has yet been built).
Going forward
More research has to be done in terms of polling and documenting the needs of the department. Furthermore, a deeper understanding about the kinds of students we’d attract and want to appoint is needed– this allows us to understand what time commitment is reasonable (both the lower and upper limits need definition). Finally, the group must from the outset be understood as something beneficial to all parties involved. The solutions named above must be deployed at launch time to ensure minimal friction, and maximum return. First steps were made last year by introducing this idea at the BIC-iGEM meeting– BIC students are excited with this idea, wherein an entire room of a dozen students raised their hands. Furthermore, there are no other groups on campus that attempt this activity, so that SOLVER would provide a unique non-conflicted service.
Of course– this all depends on the amount of work I get done on my own thesis in the first half of the semester.
Making friends
As an aside Isabelle Lam, a student I TA’d last semester in Biol-241 (microbio) has been planning on starting a job / volunteer / coop mine for science students. I should go and bother her and see how far she’s gotten. Her project is called “SPORE”. Last I checked, her team was registering a subdomain with the university.
EDIT: I previously confused Isabelle with Lisa– this has been corrected.
An Old Physiology Project — Operation Spinny Chair :D
I discovered this ancient report in my repository about three months ago– I’ve finally decided to put it up because it made my day reading the abstract again. This is definitely one of my prouder albeit sillier projects from the days of my undergrad.
Independent Research Project Acute centripetal acceleration is correlated with increased heart rate and R-wave amplitude
Matthew Boyle, Bryan Chung, Eddie Ma
Abstract
In the present study, we set out to discover the correlation between the exposure of acute centripetal acceleration in human subjects and cardiovascular function across the following three dimensions: Heart rate, R-wave amplitude and QRS interval. This was accomplished by measuring the above properties via Lead II Bipolar ECG trace, after having spun the subject at 0.8 revolutions per second in an office chair for successively, 30, 60 and 90 seconds. It was determined that heart rate showed strong positive correlation (n = 3, average increase between trials of increasing duration, 3.2 beats per minute, s = 1.8). R-wave amplitude showed positive correlation in all subjects up until and including the 60 second trial. There was no systemic correlation between duration of spin and the length of the QRS interval in any of the subjects. The heart is therefore an important effector in response to centripetal acceleration in the human model.
Key Words: electrocardiogram, QRS interval, centripetal force, R-wave amplitude, spinning office chair.
[ Download this | PDF ].Thumbnails of pages 5, 7 and 11.



Back from Conference!
Brief: The BIBM09 conference was the very first conference I have ever attended. I learned a lot from the various speakers and poster sessions–
I thought it was really interesting how the trend is to now study and manipulate large interaction pathways in silico– a theme of which is the utilization of many different data sources integrating chemical, drug and free text as well as the connection of physical protein interaction pathways and gene expression pathways. There was even a project which dealt with the alignment of pathway graphs (topology).
Dealing with pathways especially by hand and in the form of a picture is probably the bane of many biologists’ existence– I think that the solutions we’ll see in the next few years will turn this task into simple data-in-data-out software components, much like the kind we have to deal with sequence alignments.
And now, back to the real world!
Addendum: My talk went very well 🙂
And here are my slides with a preview below.


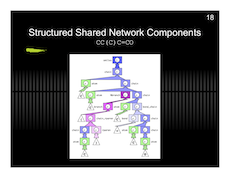
New Diagram for MSc-X3 (math paper)
Brief: I’m particularly happy with this diagram… I had something along these lines in my head for a while, but I never could figure out how to draw it correctly. I never thought that simplifying it to three easy steps was the smarter thing to do.
 Ed's Big Plans
Ed's Big Plans