Archive for the ‘Secondary Structures’ tag
TIM Barrels look like this…
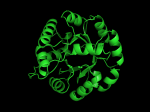
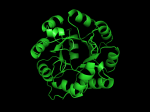
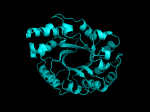
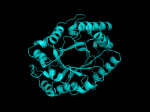
It occurred to me that I didn’t actually post any images in the past few posts. Here are two TIM Barrels that I’ve arbitrarily picked from DATE. Note– the image files are horribly misnamed, so please use the text description underneath.
From left to right– these figures are 1A53 (Beta Strand C-Face), 1A53 (Beta Strand N-Face), 1BG4 (Beta Strand N-Face), 1BG4 (Beta Strand C-Face). 1A53 is called “indole-3-glycerol phosphate synthase” and 1BG4 is called “xylanase” isolated from Penicillium simplicissimum. I won’t go into the function of each of these enzymes, but they do illustrate what a general beta-alpha TIM barrel looks like. TIM Barrels comprise of eight beta-alpha secondary structure elements. Extra helices and sheets may occur but must flank the TIM Barrel-like portion of the protein domain (1A53 has a very prominent extra alpha helix close to the camera in the far left image). The “barrel” name derives from the twisted cylinder enclosed by the parallel beta sheets in the middle of the object. TIM barrels can be deformed quite a bit too if they’re a subunit part of a larger holoprotein.
The four-character designations (1A53, 1BG4) are RCSB (A Resource for Studying Biological Macromolecules) Protein Data Bank identifiers– I’ve found that SCOP frequently links into PDB while PFAM frequently links to UniProtKB (Knowledge Base) and utilizes UniprotKB identifiers. More on that later…
 Ed's Big Plans
Ed's Big Plans