Java Classpaths are Evil
While working with Phylogenetic Analysis Library (PAL) for an alignment problem, I ran into the problem of having to specify classpaths to a jar file… it should have be straight forward enough…
Java classpaths are a pain.
Here are a few observations I’ve made about how to specify them in the command line.
- an Item can either be a directory that contains .class and .java files OR
- an Item can be a .jar file.
- to specify more than one Item.jar in Unix, use:
javac -classpath .:Item1.jar:Item2.jar
- note that you cannot put a space between the colons
- note that you must include an extra Item ‘.’ to specify the current working directory
- note that in Windows, you must use ‘;’ instead of ‘:’
- note that after compiling with javac, the same -classpath and its arguments must then be applied with java
Nuisance? Yes! Necessary Evil? No!
In the compiled Java class, there certainly could have been some metadata implemented that is a copy of the last known classpath string… why is there a disparity between the symbols used in Unix and Windows? … Why aren’t spaces allowed? Why does one have to specify the current working directory?
Evil.
A side effect of not being able to put spaces in between the colons of several paths is that one can’t just put a backslash in to negate a newline– you would need to have the next path start at the very beginning of the next line which is just ugly.
Back from Conference!
Brief: The BIBM09 conference was the very first conference I have ever attended. I learned a lot from the various speakers and poster sessions–
I thought it was really interesting how the trend is to now study and manipulate large interaction pathways in silico– a theme of which is the utilization of many different data sources integrating chemical, drug and free text as well as the connection of physical protein interaction pathways and gene expression pathways. There was even a project which dealt with the alignment of pathway graphs (topology).
Dealing with pathways especially by hand and in the form of a picture is probably the bane of many biologists’ existence– I think that the solutions we’ll see in the next few years will turn this task into simple data-in-data-out software components, much like the kind we have to deal with sequence alignments.
And now, back to the real world!
Addendum: My talk went very well 🙂
And here are my slides with a preview below.


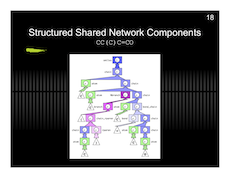
Protein Project Progress…
Last week, Liz, Aron, Andrew, Brendan and I sat down to discuss the beta-trefoil project. It was a good chance for me to understand the methods used and the kinds of results we are interested in for my own TIM Barrel project.
Continuing on with the structural repeat problem, I’ll today be writing a short FSA parser that can handle DSSP or DSS output– simply, a very primitive machine will be used to imitate a human’s visual inspection of repeated secondary structural elements in given proteins. This is in line with the work I did manually staring at structures to get a grasp of how to look at protein models, and also in line with the objective to automate much of this work. Prior to that step, I reduced the probability of doing redundant work by using BLASTCLUST and selecting only a few known structures in each cluster to inspect… a sequence based alignment for each cluster will inform me of where my manually detected repeat boundaries map to the remaining sequences.
Oddity: If you BLASTCLUST all the “FULL” (not “SEED”) sets of TIM Barrel sequences for the entire fold from PFAM along with the sequences of known TIM Barrel fold structures of SCOP, you’ll find that cluster fifteen (as of today) has these elements:
1YBE_A A6U5X9 A6WV52 Q2KDT0 Q2YNV6 Q6G0X7 Q6G5H6 Q8UIS9 Q8YEP2 Q92S49 Q98D24 A1UUA2
In the above listing, 1YBE:A (PDB code) is the sole known PDB structure, while the remainder are putative TIM Barrels (uniprot codes) as determined by the HMM model from PFAM.
The enzyme 1YBE looks like this…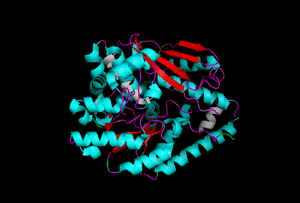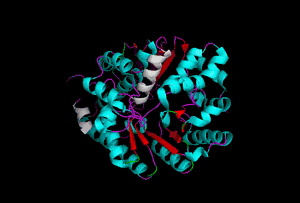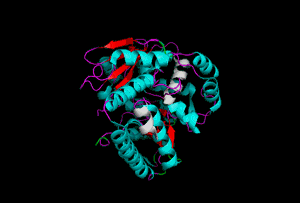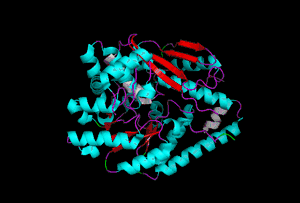
It’s an oddity because of the number of alpha-helices inserted within what is usually a hydrophobic beta barrel– the red pieces of ribbon should form a hollow cylinder, but it’s split apart for 1YBE and accommodates a bunch of cyan helices. Labeled in white are helices that break with the beta-alpha repeating secondary structural element (SSE) pattern by occurring before the first repeat. Labeled in green are breaks between beta-alpha SSE patterns.
Reference
Seetharaman, J., Swaminathan, S., Crystal Structure of a Nicotinate phosphoribosyltransferase [To be Published]
Phew! Sun Virtualbox Port Forwarding (on NAT) Solution
I finally found a solution for the port forwarding issue I was having with Sun VirtualBox (i.e. NAT provided a weird one-way mapping from which Tin (host) couldn’t dial into TinUbuntu (guest)…) — as far as I understand, this solution is guest operating system neutral but must be configured for each guest handled by a particular installation of Sun VirtualBox. That means that if you’re crazy like me and plan to put your virtual harddrives on USB sticks, you’d probably do just as well to put in a makefile that will do your configurations for you too. So here’s the solution, also saved in my wiki for extra truthiness!
Thanks to Evan and his “justwerks software” blog.
The following commands were issued…
cd /Applications/VirtualBox.app/Contents/MacOS $VBoxManage setextradata "TinUbuntu"\ "VBoxInternal/Devices/pcnet/0/LUN#0/Config/guesthttp/Protocol" TCP $VBoxManage setextradata "TinUbuntu"\ "VBoxInternal/Devices/pcnet/0/LUN#0/Config/guesthttp/GuestPort" 22 $VBoxManage setextradata "TinUbuntu"\ "VBoxInternal/Devices/pcnet/0/LUN#0/Config/guesthttp/HostPort" 2222
Which causes TinUbuntu TCP:22 to be forwarded to Tin TCP:2222.
In future, I should remove these rules and re-add them replacing “guesthttp” with my own name for this rule– In his blog, Evan has used a series of names: “guesthttp”, “guestssh” and “guestsql”– In reality, these are actually very good, unambiguous names that I’ll probably end up borrowing.
The one thing I should really dig around for is the meaning of “LUN#0”– it looks like something that’s important– the remaining virtual-directory-like objects in the configuration string look less daunting– “pcnet/0” is likely referring to the first virtual network adaptor connected to TinUbuntu.
Squishy TIM Barrel Subunits
Again with the TIM Barrel pictures! Here’s some text about it from my notes…
1a5m (A Urease) is a really interesting protein– it consists of three subunits. Each subunit consists of three unique domains: a very squashed TIM Barrel, an alpha-alpha-alpha-beta-beta domain and a beta-beta-alpha-beta-beta domain. I’m not yet sure what to call little broken alpha helices that have less than two complete turns. The TIM Barrel (though exceedingly asymmetrical) will still be accounted for in the data to be analyzed. The TIM Barrel (566 amino acids) is the alpha subunit of each symmetrical subunit. The remaining two domains are the alpha and beta subunit though PDB is not clear which is which: they each weigh in at 100 and 101 amino acids. 1a5m is part of several solved urease structures in the PDB– the collection: {1A5K, 1A5L, 1A5M, 1A5N, 1A5O} are solved by Pearson et al. (1998).
References
Matthew A. Pearson, Ruth A. Schaller, Linda Overbye Michel, P. Andrew Karplus, and, Robert P. Hausinger (1998). Chemical Rescue of Klebsiella aerogenes Urease Variants Lacking the Carbamylated-Lysine Nickel Ligand. Biochemistry. 37(17):6214-20.
Squishy squishy shapes– the giant pink object in the next picture is actually three such TIM Barrels, each of which belongs to one of the three subunits.
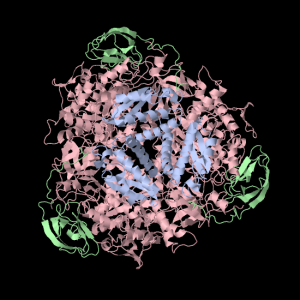
Each of the three subunits are shown separately below…
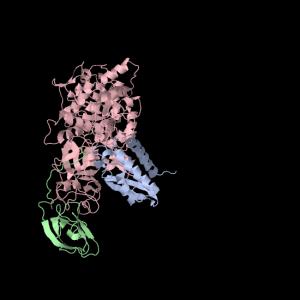 |
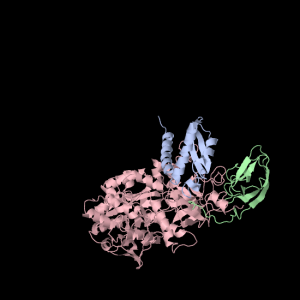 |
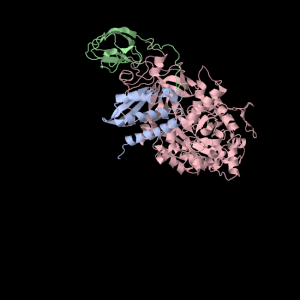 |
Aesthetically pleasing– these images were captured from the JMol output available from RCSB PDB.
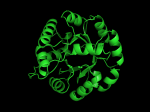
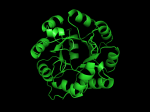
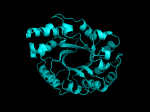
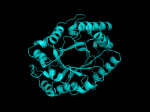
TIM Barrels look like this…
It occurred to me that I didn’t actually post any images in the past few posts. Here are two TIM Barrels that I’ve arbitrarily picked from DATE. Note– the image files are horribly misnamed, so please use the text description underneath.
From left to right– these figures are 1A53 (Beta Strand C-Face), 1A53 (Beta Strand N-Face), 1BG4 (Beta Strand N-Face), 1BG4 (Beta Strand C-Face). 1A53 is called “indole-3-glycerol phosphate synthase” and 1BG4 is called “xylanase” isolated from Penicillium simplicissimum. I won’t go into the function of each of these enzymes, but they do illustrate what a general beta-alpha TIM barrel looks like. TIM Barrels comprise of eight beta-alpha secondary structure elements. Extra helices and sheets may occur but must flank the TIM Barrel-like portion of the protein domain (1A53 has a very prominent extra alpha helix close to the camera in the far left image). The “barrel” name derives from the twisted cylinder enclosed by the parallel beta sheets in the middle of the object. TIM barrels can be deformed quite a bit too if they’re a subunit part of a larger holoprotein.
The four-character designations (1A53, 1BG4) are RCSB (A Resource for Studying Biological Macromolecules) Protein Data Bank identifiers– I’ve found that SCOP frequently links into PDB while PFAM frequently links to UniProtKB (Knowledge Base) and utilizes UniprotKB identifiers. More on that later…
 Ed's Big Plans
Ed's Big Plans